Artificial Intelligence has been doing wonders in tech-field and the induction of technology into medical science started boosting up the potential. Just in case if you don’t know, RNA, or ribonucleic acid, is present in all living cells, which acts as a messenger that dictate how proteins in the body are synthesized.
ALSO READ: Corti AI Can Detect Cardiac Arrests During Emergency Calls!
When the RNA doesn’t work properly, it can severely affect neurological, cardiovascular, and muscular regulatory processes, resulting in effects like tumors, insulin resistance, and motor skill impairment. Well, to take on that, researchers at the University of Freiburg’s Department of Computer Science developed an AI system, dubbed as LEARNA, that can learn to design RNA molecules for study.
LEARNA – Design RNA Molecules
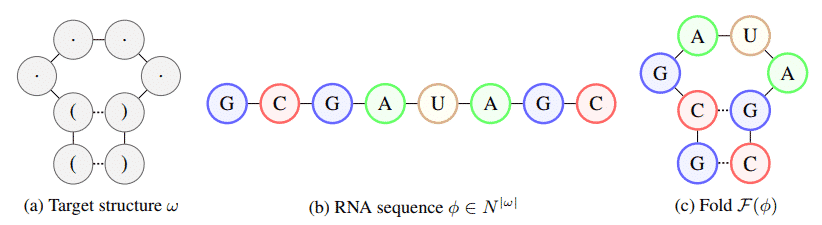
Described in a new paper (“Learning to Design RNA“) published this week on the preprint server Arxiv.org. The researcher explained the importance of RNA and also speaks about the RNA inverse folding challenge, in which they had to identify the patterns and sequences in RNA that cause it to fold into a specified structure.
Designing RNA molecules has garnered recent interest in medicine, synthetic biology, biotechnology and bioinformatics since many functional RNA molecules were shown to be involved in regulatory processes for transcription, epigenetics, and translation,” the researchers wrote. “Here, we propose a new algorithm for the RNA design problem.
The researchers’ approach relies on a deep reinforcement learning (RL) algorithm, an AI training technique that uses rewards to drive agents toward goals and also trains a policy network that can sequentially predict the entire RNA sequence. Well, this sequence folds it, and uses the distance from the resulting structure to the target structure as a signal for the AI agent.
To the best of our knowledge, this is the first application of architecture search … to RL [and] the first application of [architecture search] to metalearning.
After meta-learning across 8,000 different RNA target structure for one hour on a machine with 20 processor cores, Meta-LEARNA managed to solve up to 65 percent of the target structures in the Eterna100 benchmark (Eterna100, is a collection of 100 target structures created by players of Eterna, an online open laboratory that tasks players with creating sequences that fold to specific structures.).
Meanwhile, on another benchmark, Rfam-Taneda — Meta-LEARNA produced results as good as state-of-the-art methods in 10 seconds (compared to 90 seconds in Eterna100) and surpassed those methods in accuracy after 1 minute. The results look pretty much like those achieved by Google parent company DeepMind’s protein-folding AlphaFold system earlier this year.
Comprehensive empirical results … show that our approach achieves new state-of-the-art performance on all benchmarks while also being orders of magnitudes faster in reaching the previous state-of-the-art performance, the researchers wrote.
BONUS VIDEO
For the latest tech news, follow TechDipper on Twitter, Facebook, Google+, Instagram and subscribe to our YouTube channel.